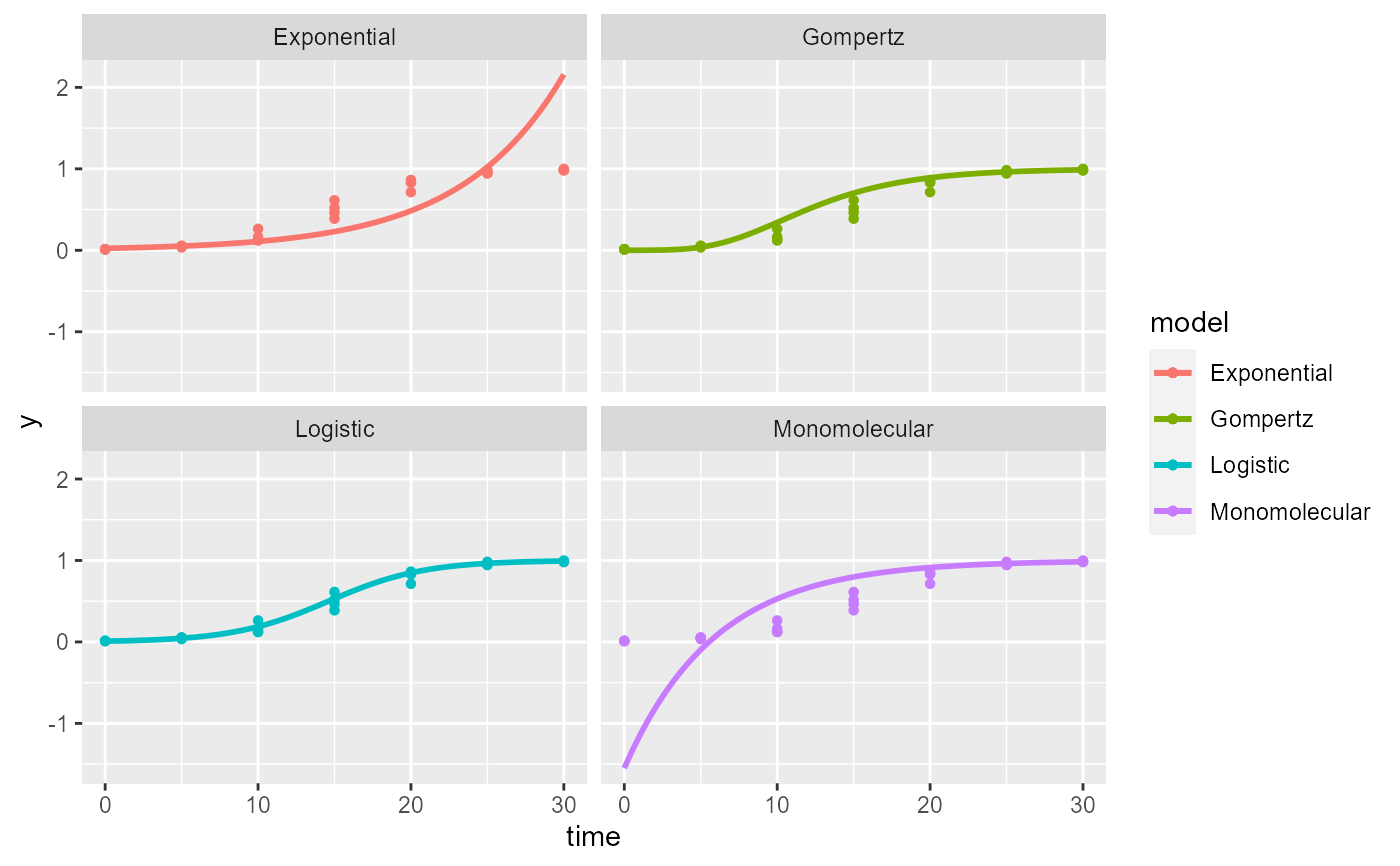
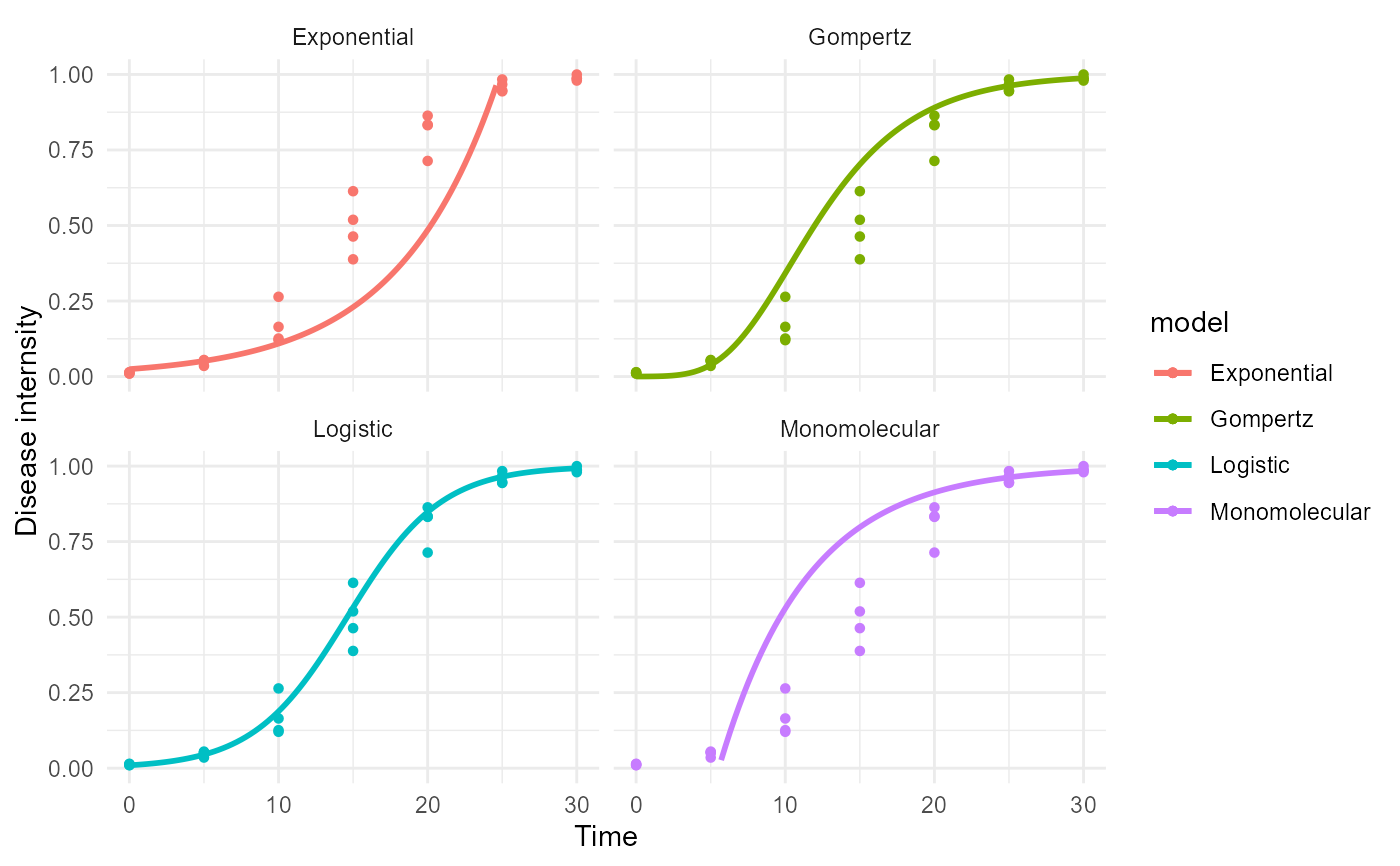
Creates a plot panel for the fitted models
plot_fit.RdCreate a ggplot2-style plot with the fitted models curves and the epidemic data.
plot_fit(object, point_size =1.2, line_size = 1, models = c("Exponential","Monomolecular", "Logistic", "Gompertz"))
Arguments
| object | A |
|---|---|
| point_size | Point size |
| line_size | Line size |
| models | Select the models to be displayed in the panel |
Details
It is possible to add more ggplot components by using the + syntax. See examples below.
Examples
epi1 <- sim_logistic(N = 30, y0 = 0.01, dt = 5, r = 0.3, alpha = 0.5, n = 4) data = data.frame(time = epi1[,2], y = epi1[,4]) fitted = fit_lin( time = data$time, y = data$y) plot_fit(fitted)# adding ggplot components library(ggplot2) plot_fit(fitted)+ theme_minimal()+ ylim(0,1)+ labs(y = "Disease internsity", x = "Time")#> Warning: Removed 18 row(s) containing missing values (geom_path).#> Warning: Removed 19 row(s) containing missing values (geom_path).